The
Sasquatch Genome Project in a Nutshell
UPDATE:
Results
of the Sasquatch Genome Study Verified by a Independent Study.
Via
Dr. Ketchum's Facebook account on 09/04/2015
HUGE NEWS: Our research has now been
independently verified genome wise. I don't know when or how they'll come out
with it and I'm not at liberty to say who yet, but it's finished and matches
what we got down to the smallest details. Thank you, God!
How it all
began
2009-2010
The TV show Destination Truth, starring Josh Gates, brought a hair sample back from Bhutan after finding a foot print
The TV show Destination Truth, starring Josh Gates, brought a hair sample back from Bhutan after finding a foot print
The
footprint:
The producers chose Dr. Melba Ketchum of DNA
Diagnostics to perform DNA analysis.
Dr. Ketchum
did not believe that a creature known as Bigfoot existed but she agreed to run
the analysis anyway. What she found astounded her. The mtDNA tested human but
the hair was not human. Dr. Ketchum remarked “The hair, visually, is not human.
It’s courser than horse tail hair” When Dr. Ketchum compared the sequence to
Genbank she could not find a match, she again remarks “There are literally
millions of sequences in this database and we’re really shocked that it didn’t
match any of the species exactly in the database”
About the
same time David Paulides, executive director of
North American Bigfoot Search, was looking for a lab to test possible
hair Bigfoot samples.
David was
having a difficult time finding a lab that would test the samples when they
found out they samples were possible Bigfoot samples. Fate brought David to
gather with Melba as the Josh Gates hair sample had her curious and now open
minded about the possibility of the Bigfoot being a real living, breathing creature.
David approached Melba to test the samples and she agreed. This began the DNA study.
My involvement
In June of 2010 I began to correspond with David Paulides
the Executive Director of North American Bigfoot Search (NABS). I realized that I needed
assistance with my research. I was encountering phenomenon and having
experiences that were extraordinary. I needed to find a support network that I
could partner with in my research. I began researching the different Bigfoot
Research organizations and I found NABS to be the most professional, respectful
and responsive group. I had read David Paulides’ book “The Hoopa Project” and
was already impressed with his logical approach to Bigfoot research, so I
introduced myself to the group via email correspondence.
I began to bounce
questions off NABS and relate my experiences to them on a regular basis. I also
began to collect hair samples and would forward them to NABS periodically. I
also sent NABS videos and photographs of the Bigfoot I was encountering in my research areas. The
relationship matured and in February of 2011 David invited me to join the NABS
Team as a Field Researcher. I gladly accepted his offer.
NABS had already
partnered with Dr. Melba Ketchum and had started a ground breaking project to
collect Bigfoot DNA specimens from all over the North American
continent.
I had started working
with NABS on this project in the spring/summer of 2010 even before I became a
team member. David and I discussed the matter and the need to humanely collect
as much DNA from the Bigfoot as possible. Through collaboration we came up
with an innovative strategy. I took clear packing tape and placed it sticky
side out. When the Bigfoot would lean or rub up against the tree their hair
would stick to the tape and be pulled out by the roots. The root of the hair
contains the DNA. I placed this “hair trap” around the tree the Bigfoot where
hiding behind when I captured them on the trail camera in August of 2010. The trail cameras were still deployed in this area on the same
trees. I was hopeful the Bigfoot would hide behind this tree again while
observing the trail camera. The effort was successful and I collected a large
amount of Bigfoot hair.
The Possible Donors:
My
Samples
In 2010 David Paulides and Melba Ketchum went on the Coast
to Coast radio show to make a public plea for Bigfoot hair and other possible
samples that could yield DNA. Hundreds of samples came in.
I sent in approximately thirty hair samples. Eleven passed
the screening process and were included in the Sasquatch Genome Study.
I took
clear packing tape and placed it sticky side out. When the Bigfoot would lean
or rub up against the tree their hair would stick to the tape and be pulled out
by the roots.
Dr. Ketchum had discovered that Bigfoot hair shafts would not yield mtDNA. The hairs must have the root or skin tag attached in order to extract usable DNA. This is a unique property of Bigfoot hair.) I placed this “hair trap” around the tree the Bigfoot where hiding behind when I captured them on the trail camera in August of 2010. The effort was successful and I collected a large amount of Bigfoot hair.
Dr. Ketchum had discovered that Bigfoot hair shafts would not yield mtDNA. The hairs must have the root or skin tag attached in order to extract usable DNA. This is a unique property of Bigfoot hair.) I placed this “hair trap” around the tree the Bigfoot where hiding behind when I captured them on the trail camera in August of 2010. The effort was successful and I collected a large amount of Bigfoot hair.
Eleven of my
hair samples went through the screening process and were tested and determined
to be from a Bigfoot.
The Team
There is a
common misconception that Dr. Ketchum worked alone with little or no
assistance. This is not true. Dr. Ketchum collaborated with at least ten other
individuals with expertise in many areas to include forensics, DNA analysis, microbiology,
molecular biology, biophysics and biochemistry just to mention a few.
Below is the
list of the team, their credentials, and their contribution to the DNA Study.
Dr. Andreas Holzenburg
Director, Professor, Department of Biology, Professor, Biochemistry and Biophysics
Director, Professor, Department of Biology, Professor, Biochemistry and Biophysics
Microscopy and Imaging Center
Texas A&M University
Has published over 30 Papers
Fan Zhang, Ph.D.
Bioinformatician in the Academic and Institutional Resources and Technology (AIRT) at the University of North Texas Health Science Center.
Dr. Pat Wojtkiewicz
Director of the Shreveport Laboratory of the North Louisiana Crime Lab System and the Technical Leader of the DNA section. He has been employed at the crime lab since 1977.
Bioinformatician in the Academic and Institutional Resources and Technology (AIRT) at the University of North Texas Health Science Center.
Dr. Pat Wojtkiewicz
Director of the Shreveport Laboratory of the North Louisiana Crime Lab System and the Technical Leader of the DNA section. He has been employed at the crime lab since 1977.
Dr. Thomas
M. Prychitko of Wayne State University in Michigan
Molecular biologist with a background that also includes evolutionary biology, microbiology and biochemistry.
Dr. Douglas G. Toler
Molecular biologist with a background that also includes evolutionary biology, microbiology and biochemistry.
Dr. Douglas G. Toler
Clinical
pathologist at Huguley Memorial Hospital in Fort Worth, Texas, for the past 30
years. His specialty also includes anatomical pathology. He is a graduate of
the University of Oklahoma Health Sciences Center.
Ms. Aliece Watts of Integrated
Forensic Laboratories in Euless, TX
BS, MS,
MT(ASCP), PBT(ASCP), F-ABC is a founding partner and the Quality Director for
Integrated Forensic Laboratories, Inc., the nation’s only private,
full-service, accredited forensic laboratory. With over 30 years of laboratory
experience and three board certifications (Forensic Biology, Medical Technology
and Phlebotomy), Ms. Watts has an extensive background in Forensic DNA and
Quality Assurance. She is an ASCLD/LAB International (ISO 17025) assessor and
has traveled to Malaysia to inspect that country’s laboratory system for
accreditation. She is a former member of the Texas Forensic Science Commission,
a Governor appointed and Senate approved position. Ms. Watts is an alumna of
the University of Texas at Arlington where she also taught Introduction to
Forensic Science Laboratory, Forensic Biology (DNA) and Methods in Forensic
Biology Laboratory.
David W. Spence - Trace evidence supervisor with
the Southwestern Institute of Forensic Sciences, Criminal Investigations
Laboratory, at Dallas County, Texas.
Sarah Bollinger, Ray Shoulders, and Ryan Smith of DNA Diagnostics.
The
following is a list of the Team and some of the areas they contributed to the
DNA Study.
Dr. Ketchum,
Dr. Pat Wojtkiecicz, David Spence, Dr. Andreas K. Holzenburg, Sarah Bollinger,
Ray Shoulders, and Ryan Smith performed experiments.
Dr. Ketchum
and Dr. Fan Zhang analyzed the genetic data.
Dr.
Ketchum., Aliece Watts, and Dr. Pat Wojtkiecicz wrote and edited the
manuscript.
Dr. Andreas
K. Holzenburg analyzed and wrote the Electron Microscopy portion of the
manuscript.
David Spence
analyzed and wrote the hair analysis portion of the manuscript.
Dr. Douglas
G. Toler analyzed and wrote the histopathology portion of the manuscript.
Aliece
Watts. also researched pertinent additions to the manuscript and helped with
data collection.
Dr. Ketchum
distributed samples, collected and combined data from the blind studies.
The Laboratories used in the
blind studies.
Again there
is a misconception that Dr. Ketchum processed all the samples in her facility.
This is not true. Dr. Ketchum realized with the implications of this study
blind testing was a must. She contracted the services of the following
laboratories to perform blind testing on the samples and return the results.
This means the laboratories had no idea what they were testing. They were just
given the samples and ask to run a DNA profile.
#
|
Laboratory
|
Type of
Testing
|
Paid
|
Blind
Study
|
Author
ship
|
|||||
1
|
North
Louisiana
Criminalistics Laboratory, Shreveport LA
|
Forensic
DNA
Extraction and DNA quantification
|
No
|
No
|
Yes
|
|||||
2
|
DNA
Diagnostics,
Nacogdoches, Texas
|
Forensic
Extraction,
Species
Screening,
Preliminary
Species
Sequencing
and STR
PP16
genotyping,
mtDNA
and nuDNA,
testing
known submitter
(human) samples.
|
No
|
Yes
|
No
|
|||||
3
|
Southwestern
Institute of Forensic Sciences, Dallas, TX
|
Hair Analysis
|
No
|
Yes
|
No
|
|||||
4
|
Family Tree DNA,
Houston, TX
|
mtDNA
confirmation
and
mtDNA whole
genome
sequencing and
haplotype
assignment,
PP16
STR confirmation
testing
and YSTR
testing.
Single locus
Amelogenin testing
|
Yes
|
No
|
Yes
|
|||||
5
|
SeqWright, Houston, TX
|
mtDNA
screening
confirmation,
sequenced
various
selected
nuDNA
loci and
confirmation
of mtDNA
species
sequencing,
sequencing
of various
amelogenin exons
|
Yes
|
No
|
Yes
|
|||||
6
|
Helix Biological
Laboratory, Department of
Biological Sciences,
Wayne State University,
Detroit, Michigan
|
Blind
parallel mtDNA
testing
of certain
submitted
samples. This
work
was done previous
to
our project and only became aware of this
testing
after we had
confirmed
mtDNA
species
ID on the same samples.
|
No
|
Yes
|
No
|
|||||
7
|
USC, Los Angeles, CA
|
Whole
genome Bead
Array SNP analysis
|
Yes
|
No
|
Yes
|
|||||
8
|
University
of Texas at
Arlington,
Wakeland
Laboratory
|
Next
Generation whole
genome
sequencing and
bioinformatics
|
Yes
|
No
|
Yes
|
|||||
9
|
UNT
Center for Human
Identification,
University
of
North Texas Health
Science
Center, Fort
Worth, TX
|
Confirmation
of
bioinformatics
|
Yes
|
Yes
|
No
|
|||||
10
|
Texas
A&M University,
College Station, TX
|
Electron
Microscopy
confirmation
|
Yes
|
Yes
|
Yes
|
|||||
11
|
University of North
Carolina, Chapel Hill,
North Carolina
|
Electron Microscopy.
The director was
unhappy about the blind
study and refused
recognition in the ms.
He did give us the data
to use.
|
No
|
No
|
Yes
|
|||||
12
|
Texas
Veterinary Medical
Diagnostic
Laboratory,
Texas
A&M University,
College Station, TX
|
Histopathology
|
Yes
|
No
|
No
|
|||||
13
|
Huguley
Pathology
Consultants,
Ft. Worth, TX
|
Histopathology
confirmation
|
No
|
Yes
|
No
|
|||||
The Samples
The screening and handling of the
hair, blood, saliva, and flesh samples.
Screening the hair
Before the
hair was considered for testing it was first screened to ensure it was not
human or any other known animal. Below
is the documentation from the DNA Study of how the hair was screened.
Hair
Analysis:
Hair samples were sent to the
Southwestern Institute of Forensic Sciences (Dallas, TX) for analysis. (Mr.
David Spence) The samples were evaluated
visually, stereoscopically, and by light microscopy to determine human or
animal origin. Hairs that were classified as potential novel hominid were also
evaluated for DNA typing potential by examining for root material. Only hairs
that were not human in appearance and could not be identified as any other
species were utilized in this study15-19.
The hairs were examined for a
variety of microscopic features such as: medulla, pigmentation, cortical fusi,
ovoid bodies, cuticle, and root and tip characteristics. The hair cuticle
patterns of select hairs were cast by embedding the hair shafts in a thin layer
of clear fingernail polish. The casts were examined microscopically.
Photographs and photomicrographs were taken of select hairs using a Canon G3
digital camera and a camera adapter (model MM3XS) from Martin Microscope.
Bigfoot hair shafts do not yield
mtDNA
During the
early stages of the study Dr. Ketchum discovered that unlike human and animal
hair Bigfoot hair will not yield mtDNA. The hair had to have a skin tag attached.
Several thousand dollars were spent on attempting to process hair shafts only.
This is why we developed a method of extracting hair that pulled them out by
the roots.
Sample Collection
The hair sample was collected from a known Bigfoot
feeding station or area of activity.
An eye witness observed the subject leave the hair sample
and the event was documented.
•
David Paulides sample from Northern California
(Raven Ulibarri – Hoopa Reservation).
•
Mary Green samples from Tennessee.
•
Janice Carter samples from Tennessee.
•
Paul Freeman sample from Washington State.
•
Robert Densford sample from Oklahoma.
•
Ericson Project samples from British Columbia,
Canada.
•
Dr. Webb Sentell sample from Louisiana.
•
Jennifer Hilaman sample from Virginia.
•
Scott Carpenter – Hair traps at feeding stations
and locations where Bigfoot had been witnesses/recorded on video.
•
Erickson Project – Samples gathered from
scratching post, blood samples obtain using sand paper on plates. (Under
supervision of a wild life biologist)
•
Dr. Al Guinn – Multiple hairs found in cast of
Bigfoot footprint.
•
Dr. Curt Nelson – Blood/Flesh from 18 inch
footprint left on nail board, Snelgrove Lake, Ontario.
Integrity
Of The Samples
•
111 Samples used in the study to include, blood,
tissue, hair, toe nail, tooth enamel.
•
17 of the samples were collected from direct
sighting locations where a witness or investigator watched Bigfoot leave
sample.
•
88 of the samples were collected by
investigators or formal research projects in and around known areas of
activity.
•
Only 6 samples were “randomly” found in the
woods where Bigfoots were known to have been active.
NO CONTAMINATION
The hair
samples that were processed were not contaminated. Critics of the study claimed
that the hair samples “had to be contaminated”. The critics were also raised
questions about of the chain of custody.
The
relevancy of the chain of custody is a fallacy. The DNA paper is a research
paper, not a criminal investigation. Where the hair came from or who handled it
is irrelevant when it concerns contamination.
From Dr.
Ketchum’s Face Book Page:
Somebody was told that the lack
of chain of custody of our samples negated our scientific findings. This is not
the case. First, most of our samples had pretty good chain of custody and some
of them were as good as any sample from a crime scene as far as documentation
and chain of custody.
Chain of custody is used in
forensics to make sure the unknown samples from the crime scene are tracked
from their recovery until they are tested and therefore not altered in any way.
This insures that nobody tampers with the samples. The samples themselves are
often of unknown origin. For example, a blood spatter on a wall could have come
from anyone, victim, suspect or even an unknown individual. Of course chain of
custody is a must when you are working a crime scene. We had really good chain
of custody on a lot of the samples. However, this testing is a little different
than a forensic case in that we were simply trying to determine the source of
the samples, human, animal, or unknown. It's nice to have chain of custody and
eyewitnesses collecting hair, but if you go out to the country and find a
strand of hair on a fence, it could be washed and tested to determine what
species it came from regardless of where it came from or who handled it because
you can't alter the DNA. We're not trying to place an individual at a crime
scene. In this type of scientific investigation, there is no crime and it
doesn't matter where it's found, only what the DNA shows matters and DNA can't
be altered with the kind of testing we did.. Therefore, it is nice but not
imperative to have chain of custody since the DNA will tell you what the sample
was from.
Decontaminating the hair samples
The DNA
study used forensic methods to wash and decontaminate the hair samples. The
process is documented in detail in the DNA study. The fact that the samples
were not contaminated is apparent in the results of the DNA analysis.
In the
Bigfoot Files - Yeti episode Dr. Sykes confirmed and validated the
decontamination protocols used by the DNA Study to ensure the hair samples were
pure and not contaminated. Below is the statement from Dr. Sykes:
The
fact is hair is very resilient. You can clean any suggestion of contamination
from the surface of the hair without damaging the DNA that lies within. This is
something we have only known in the past couple of years.
Below is the
documentation from the DNA Study of the cleaning process.
DNA
Isolation
Since
the presence of normal human DNA contamination of submitted samples was a
primary concern throughout this study, all samples were thoroughly cleaned in a
manner consistent with forensic testing procedures. In order to further rule
out contamination from human personnel and lab workers, samples from submitters
and scientists working with the samples were collected for comparison with the
results obtained in the various DNA tests.
Hair
samples were then sorted into two groups for extraction at DNA Diagnostics. DNA
from those samples containing 5-50 or more single hair roots were selected and
the roots clipped into 1.5 mL microcentrifuge tubes. The hair roots were
thoroughly cleansed with water and ethanol prior to extraction to remove any
extraneous DNA.
Flesh, Blood, Sylvia
Sample 26
- A piece of flesh, hide, and hair
reported to be from Bigfoot that was shot in Northern California.
Sample 31 -
Blood collected from a plate that had sand paper on it to cut the tongue when
the Bigfoot licked it. This was collected by the Erickson Project under the
supervision of a PhD in wildlife biology.
Sample 140 –
Blood/Plague/Tatar collected of a down spout that had been bitten into and
punctured.
Processing the samples
Sample 26 – Flesh with hair
A small
sample of flesh with hair was analyzed by Douglas G. Toler, MD, F.C.A.P. of the
Anatomic and Clinical Pathology, Huguley Pathology Consultants, P.A. 11803 S.
Freeway Ft. Worth, TX 76115. Dr. Toler determined that the skin was not human
and that the tissue was not contaminated with bacteria or fungus.
Several
hairs with follicles from Sample 26 and sent it to Southwestern Institute of
Forensic Sciences (Dallas, TX) for analysis. The hairs were processed robotically.
A core
sample from the flesh was taken to ensure no contamination. This sample
contained a high quality DNA that yielded both mtDNA and nuDNA.
The nuDNA
was of a high enough quantity and quality that core samples underwent a
structural analysis using electron microscopy.
Sample 31 Blood from food trap.
Sample 31
was collected from a research site using a food trap and forensic techniques to
ensure that there was no human contamination. The DNA extracted from the blood was of high
enough quality and quantity for a complete nuDNA genome. Sample 31 was
collected and submitted by the Erickson Project. The collect of the DNA via the
food traps was supervised by a PhD.
Sample 140 – Fresh dried blood
from down spout.
Sample 140
was fresh dried blood from the inside of a vertical downspout that had been
chewed and the individual was injured by sharp metal. The inside of the downspout
was clean and untouched by humans and the blood sample fresh and pristine.
Large bite marks, hair, and fang marks were also evident on the downspout. The
pipe was crushed to the point that the metal was torn with force. The DNA
extracted from the blood was of high enough quality and quantity for a complete
nuDNA genome.
DNA Boot Camp
What is DNA
First let’s
define DNA. DNA is short for deoxyribonucleic acid, it is the molecule that
contains the genetic code of all organisms. DNA is in each cell in the organism
and tells cells what proteins to make. A cell's proteins determine its
function. DNA is inherited by children from their parents. The DNA in a person
is a combination of the DNA from each of their parents.
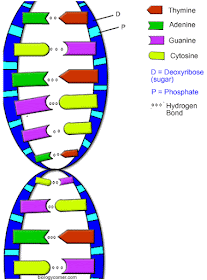
DNA has a
double helix shape, which is like a ladder twisted into a spiral. Each step of
the ladder is a pair of nucleotides. The four nucleotides in this ladder are
adenine (A), thymine (T), guanine (G), and cytosine (C). The nitrogen bases are
found in pairs. 'A' only pairs with 'T', and 'C' only pairs with 'G'. The bases
are held together by hydrogen bonds. If you unzip the DNA so you only have one
side of the ladder you still know what the corresponding nucleotide is because
DNA can only pair in this order. This is important to understand for
discussions later on in this chapter.
There are
two types of DNA, Nuclear and Mitochondrial. Mitochondrial DNA (mtDNA) is found
in cell mitochondria and is from the mother only. Nuclear DNA (nuDNA) if found
in the cell nucleus and is a combination of genetic code from both the father
and the mother. mtDNA is more abundant and easier to extract and test. nuDNA is
more difficult to extract and test therefor samples in quality and quantity are
required for successful extraction nuDNA.
Definition of terms
· Mitochondrial
DNA (mtDNA) is
found in cell mitochondria and is from the mother only.
· Nuclear
DNA (nuDNA) is found
in the cell nucleus and is a combination of genetic code from both the father
and the mother.
· Genbank is an institution that collects
and stores DNA sequences for ALL life forms on the planet. Once a DNA sequence
is placed into the database comparative analysis can be run on it using a
special software program called BLAST.
· BLAST is a software engine that
compares a DNA sequence against all the DNA sequences stored in the Genbank
database and reports any matches from this database. The current number of base
pairs stored in the Genbank database is 152,599,230,112.
The above
diagram shows the difference in mtDNA and nuDNA and from who it originates.
Now that we
have the DNA basics down, how do you perform a DNA test?
DNA is
tightly wound together in its natural state. Think of it like a jumbled wad of
rubber bands tangled and intertwined. In order to sequence the DNA the strands
must be unwound or broken into sections and unzipped. Remember you only need
one side of the helix or ladder to decode DNA because as I have previously
pointed out, A/T and C/G are always
paired together. So if we have one side of the ladder we know what the other
side has to be.
Special
enzymes are used to break down the DNA and unzip them into single strands. Once
we have the DNA broken down into strands we need to stack like strands
together. There are multiple copies of the DNA in the cell. So when the DNA is
broken down we may have thousands of copies of the same sequences. We now need
to stack these like threads together one on top of the other so we can see the
sequences. DNA testing is like a transparency picture. If you only have one
copy of the transparency it is light and difficult to see. But if you have 25
copies of the same transparency and you stack them together one on top of the
other then you can see the picture clearly. DNA testing is the same way. The
greater the DNA density the better, this will make the DNA easier to see or
what is known as “Amplifying”.
In order to “stack” the DNA together scientist use what is known as a primer. The primer’s basic function is to gather like strands of DNA together and stack them so it is easier to see the DNA sequence or amplify the sequence. In the early days of DNA testing primers had to be designed for individual species and even subspecies within a family of organisms. For example primers were developed for humans, bears, dogs, cats, pigs, cattle, etc. If you were trying to identify unknown DNA you would use the primer for the individual species on the sample. If the primer found and stacked the DNA together or amplified the DNA then you knew you had this species. If nothing was amplified then you knew you did not have this particular species.
These primers still exist today but as scientific advancements have been made in the field of DNA processing machines were developed that could process the DNA faster and more accurately. Primers were also developed to help speed up the process. What is known as a “Universal Mammal Primer” was developed. Scientist with the use of a DNA Sequencer and Universal Primers can now process DNA and get a profile faster and more accurately. Just like with the species specific primers if you use a mammal primer and the DNA does not amplify then your DNA sample is not from mammal.
Here is a very important aspect to DNA testing. The test will be affected by the quantity, quality, and purity of the DNA sample. If you have multiple copies of the DNA strands, the strands are not damaged and intact, and the DNA is all from the same organism then the primer will amplify correctly and you will get good clear results. If you have limited amounts that are contaminated the results will not amplify well. To use the transparency analogy again, now you have two transparencies of two different pictures. Some parts of the picture may be alike some different. So when the primers stack them on top of each other the picture is fuzzy and blurred and not easy to recognize.
Below is a close
up of DNA ladders on an agarose gel.
The process
I have just described in very simple language is known as Polymerase Chain
Reaction (PCR). This is the method the
independent reference labs and Dr. Ketchum used in the DNA Study to sequence
the Sasquatch DNA. Since Dr. Ketchum was aware critics and the scientific community
would be crying contamination she set up extremely strict processing methods in
order to ensure she obtained non-contaminated DNA from a single source. She
used forensic laboratories and processes to gather, wash, and then process the
DNA. The same processes and standards were used that law enforcement
laboratories use when processing DNA.
She also used forensic laboratories in blind studies so the laboratories
did not know what kind of DNA samples they were receiving. They received the
DNA, forensically processed it, and then returned the results to Dr. Ketchum.
The Testing and Results
Hair samples - Non-Human hair
yields HUMAN mitochondrial DNA
Below is the
section of the Sasquatch Genome Study Concerning the hair screening results:
The medulla and root were found to be the two most discriminating
characteristics of the microscopic examination. Most of the novel hairs had
medullary structures and diameter ratios that were clearly distinct from human
hairs. Even though a variety of medullary
structures were observed, the micrographs in Figure 5B depict those most
commonly encountered. Most of the novel hairs had elongated roots with a
somewhat “spade” shape, which is a feature of some animal hairs but is
typically not seen in human hairs (Figure 5C, left). Human hairs exhibit characteristic uniform
imbricate scale patterns of the cuticle. Several different cuticle patterns
were observed on the submitted samples. The hairs exhibited wide
imbricate scale patterns proximally that transformed to close imbricate
patterns distally. These
patterns are distinctly non-human in appearance (Figure 5C, right). Most of the submitted hairs were
not microscopically consistent with any of the hairs from the reference
collection of common animal hairs that included human, cat, dog, cow, horse,
deer, elk, antelope, moose, sheep, fox, bear, coyote, wolf, rat, mouse, monkey,
beaver, squirrel, llama and others.
The hairs were evaluated for DNA
testing by observing the presence or absence of hair roots and adherent tissue
material. Hairs with apparent translucent tissue material and/or anagen or
catagen phase roots were considered as suitable candidates for nuclear DNA
(nuDNA) testing. Hairs with telogen phase roots without tissue or hairs lacking
roots or tissue were considered suitable for mitochondrial DNA (mtDNA) testing.
Hairs that exhibited
non-human microscopic characteristics and that did not match any known animal
species were recommended for DNA testing.
The “smoking gun”
It is
important to note that until the DNA study many hair samples had been tested
throughout the years and they kept coming back “human Mitochondrial DNA”. David
Paulides noticed this trend and wrote about it on his blog
The first place that NABS turned
for information about bigfoot DNA was “The Track Record”, a newsletter produced
by Ray Crowe for 17 years. There was not an index for the track record at the
time, we reviewed all 174 documents and gleaned the DNA articles and
submissions. After reviewing the Track Record documents we turned to the web to
see what it had to offer, not much. The year we undertook this review was 2005
and many of the television shows about bigfoot did not exist.
Results
The results of our research found
that over 95% of the DNA on purported bigfoot specimens returned as “Human”.
NABS found this as an extremely puzzling result considering the “BIG” names in
the bigfoot world that consider themselves researchers proclaimed that bigfoot
was a type of ape. Many of these “researchers” claimed that the DNA showing
“human” was compromised or contaminated and the results should be disregarded.
We saw this same discussion time after time, discount the results, they are
wrong. Again, almost every moderately famous researcher known for investigating
bigfoot issues proclaimed the studies and results were flawed, every one!
Refusing to
accept the possibility that the Bigfoot could be closely related to humans the
results were always explained away as “human contamination”. In fact these were
most likely legitimate Bigfoot samples yielding human DNA. It is statically
very improbable that the scores of samples tested over the years were all
contaminated. If even one was legitimate then we have some ground breaking
findings that validate the finds of the DNA Study.
When Dr.
Ketchum analyzed the mtDNA contained in the skin tags of the Bigfoot hair every
sample tested human. The point cannot be stressed enough, NON-Human hair yielded HUMAN
mtDNA. When you link the eye witness testimony that witnessed a large
hairy bipedal hominid leaving the hair you now have empirical evidence that
Bigfoot is a human hybrid.
Below is the summary of the hair
results from the Sasquatch Genome Study:
One hundred eleven samples of
blood, tissue, hair, and other types of specimens were studied, characterized
and hypothesized to be obtained from elusive hominins in North America commonly
referred to as Sasquatch. DNA was extracted and purified from a subset of these
samples that survived rigorous screening for wildlife species identification.
Mitochondrial DNA (mtDNA) sequencing, specific genetic loci sequencing,
forensic short tandem repeat (STR) testing, whole genome single nucleotide
polymorphism (SNP) bead array analysis, and next generation whole genome
sequencing were conducted on purported Sasquatch DNA samples gathered from
various locations in North America. Additionally, histopathologic and electron
microscopic examination were performed on a large tissue sample.
The mtDNA whole genome haplotypes obtained were uniformly consistent
with modern humans. Of the 20 whole and 10 partial mitochondrial genomes
sequenced, 16 diverse haplotypes were found suggesting that these hominins did
not originate in a single geographic location.
In summary, our data indicates that the Sasquatch has human
mitochondrial DNA but possesses nuclear DNA that is a structural mosaic
consisting of human and novel non-human DNA.
Source: (http://www.human-resonance.org/sasquatch_genome.html
)
Species determination - mtDNA
The hair
samples were tested using universal mitochondrial DNA cytochrome b primers for
species determination.
The results: All
111 screened samples revealed 100% human cytochrome b. Furthermore no
heteroplasmic bases were found that would indicate contamination or a mixture. Heteroplasmy
is defined as the presence of more than one mitochondrial genome within a
tissue sample from a single individual. No heteroplasmic bases were found, the mtDNA was from a single source therefor contamination is
impossible.
mtDNA complete genome sequencing
and HV region (haplotype) sequencing results
In order for
one to understand the following results the term haplotype. A haplotype is the
group of genes that a child inherits from the mother. Like haplotypes are
grouped together and assigned a letter from alphabet. This grouping is called a
haplogroup.
HV region
sequencing will give the haplotype of the individual.
Thirty hair
samples that had ample skin tags were selected for mtDNA complete genome
sequencing and for HV region sequencing to determine the haplogroup of the
samples. The samples were sent to Family
Tree DNA for sequencing. The source of the samples was withheld from Family
Tree DNA.
Results
From the
study:
DNA samples were successfully
amplified and sequenced across the whole mitochondrial genome and the HV1 locus
using both human specific and universal primers. The sequences that were
subjected to BLAST searches in GenBank®40 showed consistent homology with human
haplotypes. No mitochondrial DNA
homology with apes, Neanderthal or Denisova cave sequences were found.
The results
showed that the mtDNA was human. The haplotypes were human. Blast searches
showed that the sequences did not match Apes, Neanderthal, or Denisovan. Of the
thirty samples twenty yielded whole mitochondrial sequences and ten yielded
partial mitochondrial genomes. The thirty samples showed sixteen different
human haplotypes.
The
interesting and unexpected finding is the origin of the haplotypes. The
majority of the haplotypes show origination from Europe, Asia, and Africa. On
only four of the haplotypes originated close to where the samples were
collected in North America.
Source for
haplotype stats and map:
The North
American haplotypes represented the following Native American Tribes:
Haplotype
A6L2c - Inuit and Nivkh peoples
Haplotype C
- Evenks and Yakuts peoples
Haplotype D
- Aleuts people
The Asian,
African, and Europe haplotypes represented the following peoples:
Haplotypes H
and T - European peoples
Haplotype V
– Arctic Saami
Haplotype
H1a (Sample 26, flesh/hair) - Basque of the Pyrenees mountains
Haplotype
L0d2a - South African Khoisan peoples
Nuclear DNA Analysis of the 30
samples
The amelogenin
gene was sequenced from the thirty samples. The amelogenin gene is
the gene in humans that is related to tooth and enamel development. It is used
in humans to determine the sex of unknown human subjects.
Results
The DNA
samples yielded four types of results: XX, XY, Y and null. The X chromosome
failing to amplify also known as “dropout” was considered by Dr. Ketchum as the
“most significant of the findings observed with the STR genotype analysis of
amelogenin.”
From the
chart above seven samples only yielded a Y chromosome. Twelve of the samples
that amplified and yielded human haplotypes failed to amplify for sex determination.
This is very strange, but in my opinion to be expected from a creature like a
Sasquatch.
Some of the
samples indicated a normal human X chromosome while others failed completely to
amplify.
When the Y
chromosome was amplified across Exons (exons are parts of DNA that are
converted into mature messenger RNA) 1, 2, 4/5 and 8, varied results were
obtained, with no samples successfully amplifying and sequencing across all
five exons (except the human controls).
When the
nuDNA sequences were compared to Genbank using a BLAST search, none of the nuDNA sequences matched any
organism in the database. Again DNA that
had human haplotypes contained sequences that do not match any known organism.
Many
sequences failed to amplify with the human primers.
From the DNA
study:
The documented anomalies came
from DNA samples that yielded long sequences with pristine electropherograms at
other loci including at least one AmelY exon. Notably, this indicated that the DNA was of high quality and that
degraded DNA was not responsible for the anomalies.
The nuDNA of
selected samples were tested for the following human specific genes:
MC1R -
human/Neanderthal red-hair color gene
TAP1 SNPs - human
antigen gene
MYH16 - This
gene is associated with the sagittal crest in apes and was analyzed in an
effort to determine if the unknown hominin was related to apes.
The Results:
MC1R - human/Neanderthal red-hair
color gene
Samples 28,
33, 35 and 37 had sufficient DNA extracted and were chosen for MC1R locus
sequencing. The primers used for the MC1R gene were for apes, humans, and Neanderthals.
The samples
were sequenced at DNA diagnostics and SeqWright lab.
DNA Diagnostic Results: The samples indicated normal
human MC1R sequence and carried alleles for red hair color in humans. The samples were not from apes or Neanderthals.
SeqWright Lab Results: Partial human sequences in some
DNA samples, while others had unknown
sequences that were not human, ape, or Neanderthal, and still others failed
to amplify.
All human
control DNA amplified and sequenced successfully from both labs.
MYH16 - This gene is associated
with the sagittal crest in apes and was analyzed in an effort to determine if
the unknown hominin was related to apes.
The primers
used were for both ape and human.
The samples
were sequenced at DNA diagnostics and SeqWright lab.
DNA Diagnostic Results: All DNA samples that successfully
amplified yielded results consistent
with humans and aligned with the human reference sequences.
SeqWright Lab Results: All DNA samples consistent with humans.
TAP1- human antigen gene associated
with breast cancer, diabetes and several other syndromes.
DNA Diagnostic Results: Some samples showed human TAP 1 sequences and others failed to
amplify. Unknown sequences were found and some amplifications with TAP 1
primers yielded long sequence of unknown origin. Two sets of 2 samples had long
unknown sequences that failed to match anything Genbank, but interestingly
matched each other. Two of these samples, samples 33 and 44, matched with one
another although there were some SNPs that were inconsistent between the two
sequences. These two samples were different in hair color and obviously were
from different individuals and were retrieved at different times. Samples 33
and 44 were collected from the same general area and the donors are thought to
be related. Samples 10 and 43 also matched each other. Samples 33/44 TAP 1
sequences were different than Samples 10/43. Sample 33 and 44 were supposed to
be related individuals, while samples 10 and 43 came from different locations.
Whole Human Genome SNP analysis:
Twenty-four
samples were tested on the whole human genome (2.5 million SNPs) Illumina® Bead
Array69 platform using the Illumina® iSCAN instrument. All of the 24 samples failed to meet the human
threshold of 95% SNP performance. The results ranged from 53% to 89% SNP
performance.
Dr. Ketchum
attempted to duplicate these odd results by purposely degrading human blood
samples. The samples were degraded with bacteria. Even though the samples were
degraded they were successfully sequenced and tested at 97.15% compared to the
89% of the non-degraded Sasquatch samples.
To further
rule out contamination and address the possibility that normal human DNA
samples could produce such markedly low SNP performance in the Illumina® Bead
Array assay, a separate group of human DNA controls was tested. These samples
were extracted from buccal swabs obtained from sample handlers and laboratory
workers and were also subjected to identical Illumina® Bead Array testing.
These human control DNA samples yielded SNP performance above the 99%.
Below is
table 8 from the Sasquatch Genome project that shows the scores of the
Sasquatch samples compared to the purposely degraded human samples.
Conclusion from the Sasquatch
Genome project:
The failures across all 2.5
million SNPs tested in the Illumina® Bead Array when compared with the human
threshold designated above 95% SNP detection, even in severely degraded human
DNA samples, suggests sequence variation from human in the actual DNA obtained
from the unknown hominin samples.
The findings reveal that
non-human hair, that yielded human
mtDNA, failed to yield human results
when the nuDNA was tested for human SNPs.
Electron microscopic examination
of Sample 26
Sample 26
was selected to be examined by an electron microscope. The sample was selected
because of its purity and lack of degradation. The electron microscope analysis
was used for the following reasons: (From the Sasquatch Genome Study)
Electron microscopic
characterization is considered superior to biochemical techniques when it comes
to an accurate topographical analysis of single- and double-stranded DNA such
as contour length/width, twisting, backfolding, nucleic acid-protein interactions,
DNA-RNA heteroduplex analysis, R-loop mapping, degradation, replication, or
packaging into higher order complexes.
The results
from the microscopic examination:
· Long double stranded DNA up to
15-20 kb, but some of the DNA also comprised smaller fragments.
· Most of the DNA showed clustering
of material along the length or at ends and were typical of disordered
single-stranded segments.
· There were also frequent
single-stranded gaps and single-stranded ends which were not observed with the
degraded human DNA control sample.
· Occasionally, with the
single-stranded ends, a back folding was observed that is typical for
unspecific base pairing that
can occur in the absence of treatment with formaldehyde glyoxal or elevated
temperatures.
· Single-stranded DNA segments
residing within double-stranded .
Summary of the microscopic
results:
Electron
micrographs of the DNA revealed unusual double strand – single strand – double
strand transitions which may have contributed to the failure to amplify during
PCR. The high quantities of single stranded DNA, interspersed with double
stranded DNA may suggest substantial structural abnormalities of the DNA
itself. The DNA was visualized twice by electron microscopy from aliquots of
two different extractions from the same sample 26. Both of these extractions
not only had yielded successful PCR amplifications previously, but were
combined and utilized for whole genome nuclear sequencing at a later time. Alternatively,
the abundance of single-stranded loci observed by electronic microscopic could
be evidence of genetic manipulation of the Sasquatch genome.
Figure 12-B
from the Sasquatch Genome Study showing the unusual double strand – single
strand – double strand transitions.
Below is a
graphic drawing of the unusual double strand – single strand – double strand
transitions.
Next Generation Whole Genome
Sequencing
Due to the large
number of failures to amplify, failure to meet the human SNP threshold and
unusual sequences obtained, Dr. Ketchum selected three samples (26, 31 and 140)
with large amounts of high quality DNA were selected for next generation whole
genome sequencing.
The DNA from
these three samples was sequenced using the next generation Illumina platform
at the University of Texas, Southwestern in Dallas, TX, a laboratory that
sequences human genomes.
In depth
analysis of all three genomic sequences (samples 26, 31 and 140) was performed
at the University of Texas, Southwestern and alignment confirmed by the
University of North Texas Health Science Center.
Q30 Scores
The run
summary generated by the HiSeq 2000 next generation sequencer provides scores
called Q30 for run quality and purity of the samples. The Q30 can also be used
to determine if there was any contamination (or mixture) found in the samples
sequenced. According to Illumina, a pure, single source sample would have an
Q30 score of 80 or greater with an average of 85. However, if there was contamination
present in the sample sequenced, the sequences would compete against one
another prior to sequencing causing a contaminated sample to have a Q30 score
of 40 to 50. The Q30 scores for the three genomes sequenced had Q30 scores of 88.6, 88.4 and 88.7 for samples 26, 31
and 140. The Q30 scores conclusively prove that the sequences were from a
single source, non-contaminated, and that the qualities of the sequences were
far above the average genome (80) sequenced using the Illumina next generation
sequencing platform.
Results
- The three genomes matched each other and were form the same “unknown” species.
- The three genomes were compared to all the genomes in Genbank. The BLAST search did not match any known sequence in the Genbank database.
- The mtDNA sequences produced by the Illumina next generation platform matched the mtDNA sequences extracted from the same samples by Family Tree DNA in the early stages of the study.
- The genomes from samples 26 and 140 were used to produce a primate “family tree” using a tool provided by Genbank called BLASTn. This produces a graphic representation of where the genome falls in the primate family.
Summary
Over a five year period over
one hundred samples of hair, tissue,
tree bark shavings, saliva, and blood submitted by a nationwide team of
submitters from 14 US states and two Canadian provinces.
All samples were screened prior
to DNA analysis to eliminate common wildlife species using a variety of
methodologies including microscopic morphological examination and comparison
against a large reference collection of known North American wildlife hair.
Over one hundred hair samples
passed the initial screen and were determined not to be human or from a known
wildlife species.
It was discovered during
testing that Sasquatch hair alone would not yield mtDNA. This was unusual
because both human and animal hair will yield mtDNA. Only hair samples with
ample skin tags were selected could be used for the study.
To avoid contamination the hair
samples were forensically processed and cleaned.
Sylvia, flesh, blood, tooth,
finger nail and enable samples were also submitted to the project.
The hair samples were sent to
multiple labs for blind testing. All the samples were determined to be human,
with results consistent with human mtDNA.
Non-human hair yielded human mtDNA.
Results from purported Sasquatch
samples tested over the previous two decades had always yielded a result of
human mtDNA. These results were always dismissed as “human contamination”. The
results of the Sasquatch DNA study support the conclusion that these samples
were in fact Sasquatch samples.
Thirty
mtDNA samples were selected for complete genome sequencing and HV region (haplotype) sequencing
results. The samples yielded haplotypes that showed the origination of the Sasquatches
to be from Asian,
African, and Europe. Only four out of
the thirty haplotypes originated close to where the hair samples were collected
in North America. Also none of the
samples contained mtDNA from apes, Neanderthals, or Denisovians.
The
amelognein gene from the nuDNA of the same thirty samples was sequenced to
determine the sex of the subjects. Mixed results were obtained for human
chromosomes to others failing to amplify, while others would only amplify the Y
chromosome. Again we pure, non-contaminated samples that yielded human mtDNA
failing at times to amplify when human primers are used for sex determination.
The samples
were tested for the MC1R - human/Neanderthal red-hair color gene, MYH16 - This
gene is associated with the sagittal crest in apes and was analyzed in an
effort to determine if the unknown hominin was related to apes, and the TAP1-
human antigen gene associated with breast cancer, diabetes and several other
syndromes. The MC1R results were human, no ape or Neanderthal sequences. The MYH16 results were human. The TAP1
results showed unknown when compared to the Genbank database, but the samples
matched each other.
Twenty four
samples were tested on the whole human genome (2.5 million SNPs) Illumina® Bead
Array69 platform using the Illumina® iSCAN instrument. All of the 24 samples failed to meet the human
threshold of 95% SNP performance. The results ranged from 53% to 89% SNP
performance. Attempts were made to
duplicate these results with degraded human control DNA. The attempt was
unsuccessful. The degraded human DNA scored 97.15% compared to the non-degraded
Sasquatch DNA that scored 89% at best. The
mtDNA from these samples tested as human but the nuDNA results for the SNPs
were non-human.
A core sample was taken from the flesh sample
number 26 and examined under an electron microscope. The results showed the
nuDNA to be very unusual and non-human. Electron micrographs of the DNA
revealed unusual double strand – single strand – double strand transitions. This could suggest evidence that the
Sasquatch nuDNA had sometime in the past been genetically manipulated.
Three
samples that contained ample nuDNA were selected for Next Generation Whole
Genome Sequencing. The DNA from these three samples was sequenced using the
next generation Illumina platform at the University of Texas, Southwestern in
Dallas, TX. In depth analysis of all three genomic sequences (samples 26, 31
and 140) was performed at the University of Texas, Southwestern and alignment
confirmed by the University of North Texas Health Science Center. The resulting
genomes were compared to the complete Genbank data base using a BLAST search. No match was found the three genomes
were from a completely unknown species yet the three samples aligned and
matched each other.
Conclusion:
DNA from
over one hundred separate samples of hair, tissue, toenail, bark scrapings,
saliva and blood obtained from dozens of collection sites throughout North
America have been extracted, analyzed and sequenced. The hair samples were not
consistent with human or any known wildlife hairs. Analysis of the DNA found
that the mitochondrial DNA was human, but the nuclear DNA was from a unknown
source. All known ape and relic hominin species such as Neanderthal and
Denisovan were eliminated as being contributors to both the nuclear and
mitochondrial sequences. Whole genome analysis
showed that the Sasquatch is an unknown species with some DNA sequences not
matching humans or any known species while other sequences matched humans and
other primates. The conclusion of the Sasquatch genome study is as follows:
The data clearly supports that
these hominins exist as a novel species of primate. The data further suggests
that they are human hybrids originating from human females. This hybridization
can be likened to humans with Denisovan admixture resulting from Denisovan
males mating with human females103. The same type of mating potentially
occurred with Sasquatch; however, in the case of Sasquatch, the admixture is
human. Though preliminary analysis supports the hybridization hypothesis,
alternatively, it could also be hypothesized that the Sasquatch are human in
origin, having been isolated in closed breeding populations for thousands of
years. Nevertheless, the data
conclusively proves that the Sasquatch exist as an extant hominin and are a
direct maternal descendent of modern humans.
The Sasquatch is a human hybrid
species, the mtDNA is human and the nuDNA is unknown.
Zoobank
Registration of the Sasquatch
Homo
Sapiens Cognatus - Cognatus is from the Latin which essentially means “related
by blood.”
External Support for the Sasquatch
Genome Study
Given the
toxic nature of this subject most mainstream scientist have stayed away from
the paper, afraid for their careers. The biased against the study is
institutional and wide ranging. Though it is a subject for another book the Sasquatch
Genome Study was caught in the crossfire in the war between the evolutionist
and the creationist. Darwin’s theory is dying in my opinion but absent any
cogent theory the Darwinist (99% of mainstream science) must protect the theory
at any cost. Since the Sasquatch Genome Study could call into question many of
the tenants of the Darwinist it had to be squashed.
There was
also an attitude pervasive in the mainstream that just could not allow “A woman
veterinarian from Texas and a bunch of rednecks” to make one of the most
profound discoveries of the century. Below is a quote from Dr. Kethcum:
"It seems mainstream science
just can’t seem to tolerate something controversial, especially from a group of
primarily forensic scientists and not “famous academians” aligned with large
universities, even though most of our sequencing and analysis was performed at
just such facilities.
We encountered the worst
scientific bias in the peer review process in recent history. I am calling it
the “Galileo Effect”. Several journals wouldn’t even read our manuscript when
we sent them a pre-submission inquiry."
There has
been support for the study amongst a quiet but growing group of scientist and researchers.
One of the
supporters is Dr. David Swenson, he is a Biochemist and has over 39 journal
publications to his credit. He looked at the paper and the data and made the
following statement:
Brien Foerster, Jeff Kart, and
other interested parties. I went over the manuscript by Melba Ketchum on
Bigfoot genomics. My desktop had difficulty with a blast analysis of the
consensus sequences. It helped me understand more about the project. This
collaborative venture has done a huge project that taxes me to fully grasp. I
see interesting homology with a standard human sequence with 99% match for
mitochondria. From my abbreviated study, the nuclear genome seems to have human
and nonhuman sequences.
My opinion of the creature is
that it is a hybrid of a human mother and an unknown hominid male, Just as
reported. For all practical purposes, it should be treated as human and
protected under law.
Brien, selection of Melba's lab
for your studies is a very good call.
Sasquatch is real, as proven by
genetic analysis.
A study
published in the Journal Nature appears to support the general findings of the
DNA study. One of the main criticisms of the Sasquatch DNA Paper published in
early 2013 was that hybridization between Sasquatch and modern humans was
"ludicrous", "impossible", or "wild speculation".
But wait, fast forward to December of 2013 and a group of scientist publishes a
paper in the Journal Nature that essentially claims that the genome of a
Neanderthal skeleton they discovered was actually a hybrid, a mixture of modern
humans, Denisovans, Neanderthals, and a "potential unknown hominin".
From the
Journal Nature study:
"The new analysis finds that
the genomes of Han Chinese and other mainland Asian populations, as well as of
Native Americans, contain about 0.2 percent Denisovan genes…..
The genome comparisons also show that
Denisovans interbred with a mysterious, fourth group of early humans also
living in Eurasia at the time."
Sources:
(http://www.sci-news.com/othersciences/anthropology/science-neanderthal-genome-fourth-lineage-01624.html) and (http://www.nature.com/nature/journal/v505/n7481/full/nature12886.html)
The
results are extremely similar to Dr. Ketchum's results, a genome that shows
hybridization with modern humans and a unknown hominin.
A world renowned genetic
scientist Dr. Tom Gilbert, of the University of Copenhangen, recently sequenced
hairs from a purported Orang Pendek (A Sasquatch like creature that lives in
South East Asia).
He received hairs that were
analyzed and determined not to be human or from a known animal. When he tested
the mtDNA from these non-human hairs the results came back human.
Though critics claim
contamination, Dr. Gilbert is a world renowned Professor of Genetics and I
would assume he would know how to properly process a DNA sample without
contaminating it.
Dr. Gilbert has over 125
peer reviewed publications to his credit. He recently was lead author of the
paper where the whole genome was sequenced from a 12000 year old Native
American child (Anzick-1).
YouTube video link - https://www.youtube.com/watch?v=UcPrXZyBUfo
Gofundme
support
From Dr. Ketchum:
Hi all, just wanted everyone to
know we do have academics supporting the paper and our research via the
Gofundme campaign. These two folks were kind enough to donate to the sequencing
project and put their names on their donations. I just thought you would like
to see that we do indeed have support from other scientists and these guys
actually put their money down to prove it! I've posted them below but you can
go to the page here to see for yourself: http://www.gofundme.com/b1f5ak
Libby
Rutledge, Ph.D. - $100
Thank you so much for your Sasquatch Genome Project paper. I assign this paper for discussion in my senior molecular biology class at Salish Kootenai College.
Thank you so much for your Sasquatch Genome Project paper. I assign this paper for discussion in my senior molecular biology class at Salish Kootenai College.
Dr.
Philip Haseley - $150
This is a challenge to all the wimpy academic types to come forward and support this research likewise.
This is a challenge to all the wimpy academic types to come forward and support this research likewise.
George Knapp
December 2012
Scores of hair samples were sent
to a dozen well-respected DNA labs across the country. The people at the labs
weren’t told anything about the samples. They performed DNA analysis in the
blind, then sent the remarkable findings back to Ketchum. I’ll put it this way
— this is spooky stuff. The results are unequivocal: The hairs are not only
from an unknown species, but they show a common link to humans. In other words,
whatever these creatures are, they share a common ancestry with humans dating
back about 15,000 years. Half of the DNA in the samples is simply unknown. The
blind tests conducted by various labs weeded out known species such as bears or
wolves. And in the end, they were left with the completely uncomfortable
conclusion that the hairs came from a primate species previously unknown to
science.
Source: (Knapp, George – Las
Vegas City Life, December 2012)
The
Contamination Myth
Mainstream scientific critics of
the study have been relying on their old standby “the samples were contaminated
or not handled properly”. This claim is refuted throughout the paper beginning
the forensic washing of the hairs that even Bryan Sykes admits “You can clean
any suggestion of contamination from the surface of the hair without damaging
the DNA that lies within. This is something we have only known in the past
couple of years.” Dr. Ketchum knew before starting the study this would both a
concern and a criticism so throughout the study steps were taken to ensure both
no contamination and quality samples. The results themselves prove there was no
contamination. This is demonstrated by the high Q30 scores to the lack
heteroplasmic bases when the samples
were screened for cytochrome b.
Even those who had openly criticized
the Sasquatch genome study and made unsubstantiated personal attacks toward Dr.
Ketchum and other members of the team admitted there was no contamination. Below
is an excerpt from Robert Lindsay’s blog:
"No contamination in Ketchum
DNA findings. There is some little-known evidence that there is no
contamination in her samples: Ketchum tested the Bigfoot nuDNA for several
human genes, the names of which you can find in the manuscript. MC1R
(human/Neanderthal red-hair color gene) showed up in the Bigfoot nuDNA as did
the human antigen gene TAP1 (most of the time) and the jaw muscle gene MYH16
(which when present showed only a human profile rather than an ape one).
Not discussed in the manuscript
are the tests Ketchum did for the TYR gene, which is associated with skin
pigmentation, and the HAR1 gene, which is a “human accelerated region”
associated with human neurological development. The human skin color gene TYR
and human brain gene HAR1 were not found in Bigfoot nuDNA. Now that in and of
itself is very interesting.
If the samples really were just
bear or coyote or bobcat smeared with human contamination, all of the human
genes should show up all over the place. The peer-reviewers for Ketchum’s
manuscript only wanted positive, not negative, results included for gene tests,
so the TYR and HAR1 data are not discussed in the manuscript. However, you can
see the remnants of it in the Supplemental Data 12 appendix. The bottom line is
the Bigfoot nuDNA is missing some important human genes that should be there if
the nuDNA were in fact simply contaminated with human DNA.
Furthermore, if the samples were
simply bears, coyotes or whatever with no human contamination present, the
human genes listed above would not be there at all.
The conclusion is that the
“contamination” meme bandied about is simply a red herring. Ketchum’s DNA
results, whatever they were and whatever they mean, there simply not a result
of contamination in any way, shape or form. Critics really need to get over the
contamination BS."
(Source: http://robertlindsay.wordpress.com/ )
Results of the Sasquatch Genome
Study Verified by an Independent Study:
Via Dr. Ketchum's Facebook
account on 09/04/2015
HUGE NEWS: Our research has now been
independently verified genome wise. I don't know when or how they'll come out
with it and I'm not at liberty to say who yet, but it's finished and matches
what we got down to the smallest details. Thank you, God!
The
results from my DNA samples
Sample #2 – Collected July 2011 –
Brown hair
Haplotype – T2b
Same as samples (1,12,36)
Bigfoot in 1 = Oklahoma, 12 =
Oregon, 36 = Washington
Amelogenin – XX (Male)
Sample #3 – Collected July 2011
Amelogenin – XY (Female)
SNP – 62.25
Sample #42 – Collected February
2011 – Reddish brown to brown
Haplotype – T2
Same as samples (39b, 41)
Bigfoot in 39b = Southern Alaska,
41 = North Texas
Amelogenin Seq Results
Only Exon 2 – H
Sample #11 – Collected August
2009 – Brown hair
Haplotype – A6L2c
Amelogenin – XY (Female)
Sample 38 – Collected October
2010 – Curly brown hair
Haplotype – V2
Amelogenin Seq Results
Exon 2,4/5,8 – H
SNP – 64.21
Sample 82 – Collected May 2011 –
Light brown to blonde
Amelogenin Seq Results
Amelx, 8 – H
Sample 122 – Collected May 2011 –
Mixed color
Amelogenin Seq Results
Amelx=H, 2=H-Chromose 13, 4/5 = H
Click here to view/download the Sasquatch Genome Study








































Very good account of the developments so far. Sadly you do not mention that some other independent labs - according to Ketchum`s facebook page - have examined her study`s results in the meantime and one of them informed Ketchum two weeks ago that they indeed have had the same results as her. Let`s hope that the (alleged) lab will publish their findings soon and hopefully will "whitewash" Ketchum and her project.
ReplyDeleteAdded the update, oversight on my part, thanks!
DeleteI wish I had a better science background to say how I feel about other
ReplyDeletelabs confirming Dr. Ketchum's results. But all I say is to quote our
favorite US Marine Gomer Pyle, " Surprise, Surprise, Surprise ".
Some people are more imaginative, less anxious, more broad minded, have fewer vested interests in fixed paradigms, and can see what is actually before them, without the influence of ego, without fear or favor.
ReplyDelete